8.5 Mating types in Basidiomycota
The breeding system in Basidiomycota relies on two MAT loci. One locus encodes tightly linked Pheromones and pheromone Receptors (now called the P/R locus), and the other encodes homeodomain-type transcription factors (called the HD locus), which determine events following syngamy. In basidiomycetes, as in other filamentous fungi, syngamy means ‘hyphal fusion or anastomosis’ as described in Section 5.16, above). For a successful mating that completes the sexual cycle and produces progeny spores, the two haploid hyphae that fuse must differ at both MAT loci.
When the two MAT loci are on different chromosomes, meiosis can generate four mating types among the haploid progeny because the meiotic nucleus must be heterozygous at both MAT loci. The occurrence of four progeny mating types defines this as a tetrapolar breeding system. Some basidiomycetes have a bipolar breeding system instead, which is controlled by a single MAT locus because either the P/R and HD loci are closely linked on the same chromosome and segregate together, or because one has lost its function in determining mating-type. About 65% of the species in the Agaricomycotina (see Section 3.8) are tetrapolar, whereas Ustilaginomycotina and the Pucciniomycotina are predominantly bipolar. Agaricomycetes, which includes most fungi that produce conspicuous fruit bodies, have evolved a system to increase the diversity of ‘alleles’ at both MAT loci, in some cases yielding species with hundreds or thousands of possible mating types. Tetrapolar breeding systems, and especially this great diversity in MAT ‘alleles’, are believed to be adaptations to promote outbreeding in organisms that, for the most part, rely on air currents to distribute their spores (Section 8.6, below).
The P/R system of Basidiomycota that governs hyphal fusion depends on small (10- to 15-amino acid) lipopeptide pheromones, which are made from precursor polypeptides containing 35 to 40 amino acids by post-translation modifications at both the N- and C-termini (Raudaskoski & Kothe, 2010; Raudaskoski, 2015). All active basidiomycete mating pheromones isolated so far are hydrophobic diffusible lipopeptides that undergo farnesylation at the C-terminal cysteine residue of a CAAX motif (‘C’ is cysteine, ‘A’ an aliphatic amino acid, and ‘X’ is any residue) and amino-terminal processing. These diffusible pheromones are received by pheromone receptors, located in the plasma membrane with seven transmembrane domains, which are coupled to a G-protein signal transduction cascades. This has been investigated in greatest detail in Ustilago maydis, where two interconnected signalling pathways, one involving cAMP-dependent protein kinase and the other a MAP kinase (Section 5.13, above). The two pathways converge to a HMG-transcription factor (Section 8.1, above) called Pheromone response factor (Prf1) which recognises and binds to the specific ‘pheromone response motifs located in the regulatory regions of the genes that the pheromone induces. Although the protein kinase cascade pathways are conserved over broad evolutionary distances, the transcription factors that ultimately activate or repress the specific target genes are often species-specific (Coelho et al., 2017).
This way of controlling mating fusion is similar to the a- and α-factor P/R system of ascomycetes and is thought to predate the separation of Asco- and Basidiomycota clades. It is feasible that what is common in this shared P/R system contributes to the information interchange that takes place between the two fusing hyphal cells involved in fusion we described in Section 5.16, above. However, there are important differences between ascomycetes and basidiomycetes, in that only homologues of the a-factor pheromone and Ste3 pheromone receptor of the ascomycete system exist in basidiomycetes.
Identity-sensing specificity in basidiomycetes is done by allelic variants of the same type of genes, rather than depending on the ‘a-factor/Ste3’ plus ‘α-factor/Ste2’ coupled sensing system characteristic of the ascomycetes (Casselton & Olesnicky, 1998; Kües, 2015). Mating (syngamy or fusion) is initiated by a reciprocal exchange of pheromones recognised by matching pheromone receptor variants in both mating types, and thus the two mycelia involved need to carry different alleles of the pheromone and receptor genes at the P/R locus.
Many additional pheromone receptor-like genes have been identified in Agaricomycetes that are non-mating-type-specific receptors and are not sufficient to induce mating-specific development. Many of these nonmating-type-specific receptors are located close to the P/R locus, while others are unlinked. Three distinctive features of these receptors are that:
- (i) they have longer C-terminal cytoplasmic regions,
- (ii) they lack pheromone genes in close association, and
- (iii) they show lower levels of intraspecific polymorphism.
Non-mating-type-specific receptors have been found in Coprinopsis cinerea, Phanerochaete chrysosporium, Laccaria bicolor, Postia placenta, and several polypores (Coelho et al., 2017). Possible functions include a role in monokaryotic fruiting or in communication within the vegetative mycelium.
The HD locus encodes a pair of homeodomain proteins, HD1 and HD2. Their open reading frames are normally adjacent and divergently transcribed. Together these provide what has been called the second compatibility checkpoint (Coelho et al., 2017). Compatibility of the genetic components at the HD locus is required to produce viable progeny from basidiomycete matings. What happens after the P/R system has arranged successful hyphal fusion (syngamy) depends upon the formation of functional heterodimeric transcription factors in which dimerisation is limited to HD1 and HD2 proteins that originate from two haploid nuclei that have different alleles at the HD locus. This requirement ensures that active heterodimers are only formed in dikaryotic hyphal cells and do not arise in the haploid (monokaryotic) hyphal cells of either parent.
Dimerisation between HD1 and HD2 proteins involve polar-to-hydrophobic interactions with allele-variable cohesive contact interfaces contributing to binding affinity. HD1 and HD2 proteins from the same allele are unable to form heterodimers because mismatches in their amino acid sequences prevent cohesion in the monokaryon. The interfaces involved in these HD1-HD2 interactions that determine allele specificity are located on the N-terminal side of the DNA binding motifs. This is the region that is highly variable between different alleles of both HD1 and HD2 open reading frames. Once formed, the functional heterodimers will bind to promoter sequences of their target genes leading to the induction of the specific sets of genes involved in subsequent differentiation (Bobola & Merabet, 2017; Vonk & Ohm, 2018). This differentiation varies according to the life styles of different fungi and includes:
- a morphological switch from yeast-like cells to filamentous growth and development of pathogenicity, as in Ustilago maydis and other smuts (Pérez-Martín & de Sena-Tomás, 2011);
- vigorous vegetative growth of the dikaryon followed by the ability to form the fruit body in the mushrooms and related fungi (Kruzel & Hull, 2010; Kües, 2015; Coelho et al., 2017).
The sequences around the HD locus are generally conserved in Agaricomycetes. Two genes usually border this locus: one encodes a Mitochondrial Intermediate Peptidase (MIP), and the other is known as a β-flanking gene (β-fg) for an unknown protein. Although this configuration is found in most species analysed, exceptions have been reported. In Schizophyllum commune, for example, two large rearrangements at the HD locus have taken place causing the separation of the HD locus into two clearly distinct subloci that now lie more than 500 kb apart and frequently recombine during meiosis. A similar situation is observed in Flammulina velutipes, but the distance between the two HD subloci is about 70 kb. Other exceptions to the typical MIP-HD1/HD2-β-fg gene arrangement were observed in the cacao pathogen Moniliophthora roreri, where both MIP and β-fg are located 40 and 60 kb upstream of HD1/HD2, respectively, and in Lentinula edodes, where MIP is distant from the HD locus. Gene order conservation at the P/R locus is also observed among Agaricomycetes, although to a lesser extent compared to the HD locus. There is a single mating-type locus in the commercial button mushroom Agaricus bisporus var. bisporus that corresponds to the HD locus (flanked by MIP and βfg), the PR locus having lost its mating-type role. Recombination is suppressed across whole chromosomes in A. bisporus var. bisporus, but suppression of recombination at the MIP gene is caused by a 140 kb-long inversion between mating types in an A. bisporus var. burnettii heterokaryon (Foulongne-Oriol et al., 2021). The P/R locus in Laccaria bicolor is in a chromosomal region prone to gene duplication, translocations, and accumulation of transposable elements, and in the cantharelloid wood-decay basidiomycete Botryobasidium botryosum, there are transposons in the immediate vicinity of pheromone and receptor genes.
Ustilago maydis causes the smut disease of maize. It has a tetrapolar mating system comprising one 'multiallelic' mating type factor and one with only two 'alleles'. Ustilago produces unicellular, haploid sporidia that grow vegetatively by budding like a yeast phase; these can be cultured on synthetic media and are non-pathogenic for the host plant (CLICK HERE to see Fig. 14 in Chapter 3).
Conjugation tubes are formed when sporidia of opposite mating type are mixed, and fusion of these produces the dikaryon, which then grows as a filamentous fungus. The dikaryon is the pathogenic stage. Fusion of sporidia is controlled by the biallelic ‘a’ mating type locus, the heterozygous a1/a2 genotype being required for conjugation and the transition between the yeast and filamentous forms. The multiallelic ‘b’ locus stops diploid cells fusing, determines the true hyphal growth form, and pathogenicity (Fig. 7).
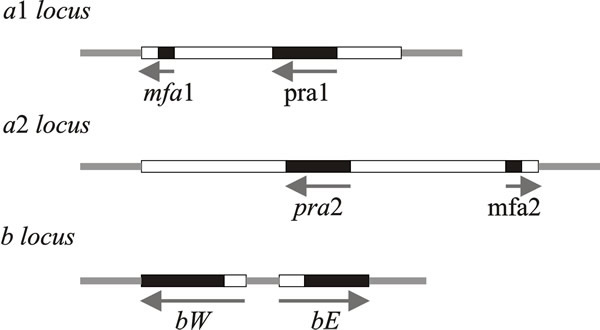 |
| Fig. 7. Schematic representations of the structures of the a and b mating type loci of Ustilago maydis. Idiomorphs of the a locus consist of mating-type specific (i.e. variable) DNA sequences (4 500 base pairs in a1, 8 000 base pairs in a2), here shown as open boxes, within which are the genes for mating (mfa and pra). The b locus has two reading frames, bW and bE, which produce polypeptides containing domains of more than 90% sequence identity (shown as black boxes) and variable domains (open boxes) which show 60 to 90% identity. Arrows indicate the direction of transcription. The PRE sequence mentioned in the text is a very short control site upstream of the pheromone genes, mfa1 and mfa2. Modified from Chapter 2 in Moore & Novak Frazer, 2002. |
The a1 idiomorph consists of 4.5 kb of DNA and the a2 idiomorph is 8 kb. Two genes have been identified in these regions: mfa1 (in a1) and mfa2 (in a2) code for pheromones, and pra1/pra2 encode pheromone receptors. The pheromones diffuse away from their producer cells and induce conjugation tubes after binding to pheromone receptors on cells of opposite mating type.
Pheromone signalling is also necessary for the maintenance of the filamentous dikaryon after cell fusion. Pheromone induces all of the mating-type genes to levels 10 to 50 times higher than their basal level. The upstream control element responsible for this pheromone stimulation, which is called the pheromone response element (PRE), has the sequence ACAAAGGG. It is on the same DNA molecule as the pheromone genes so it is called a ‘cis-acting element’, by analogy with chemical terminology which describes two substituents on the same side of an axis of symmetry in a molecule as the cis-configuration, and the alternative of two substituents on opposite sides of an axis as the trans-configuration.
The PRE sequence is similar to the consensus sequence recognised by HMG polypeptides, including MAT a-1 of N. crassa (see above). A gene called pfr1 (pheromone response factor), the product of which binds to the PRE sequences found in both a1 and a2 idiomorphs, encodes the controlling transcription factor. The downstream pheromone response pathway includes at least one MAP kinase encoded by a gene called fuz7, which is homologous to the ‘archetypal’ MAP kinase gene of Saccharomyces cerevisiae, STE7 (a MAP kinase is a ‘mitogen activated protein kinase’, where a mitogen is any agent that induces mitosis). Disruption of fuz7 results in phenotypes that show that fuz7 is involved in a-dependent mating events like conjugation tube formation, conjugation and establishment and maintenance of filamentous growth. Other components of the pheromone response pathway that have been found are four genes encoding G-proteins (gpa1 to 4). There are indications that fuz7 and gpa3 do not belong to the same pathway, so there may be several pheromone responses in U. maydis, either in parallel or in series.
The b mating type factor contains two genes which are transcribed in opposite directions (Fig. 7): bE and bW (= East and West) with coding sequences equivalent to polypeptides of 473 and 629 amino acids, respectively. The amino terminal end of the coding sequence is highly variable, whereas the carboxy-terminal end is conserved in different b idiomorphs. The bE and bW proteins are, respectively, the HD1 and HD2 homeodomain proteins that form a dimer that is a transcription activator. This activates transcription of genes required for the sexual cycle and/or repression of haploid-specific genes (interaction of the equivalent Coprinopsis homeodomain proteins is illustrated in Fig. 10 below).
Dimers comprised of bE & bW from the same idiomorph are inactive; the heterodimer functions properly only when the proteins come from different idiomorphs. The proteins encoded by these genes contain sequences homologous with DNA-binding homeodomain regions of known transcription-regulating factors, which is why they are called HD1 and HD2. The homeodomain is an extended helix-turn-helix DNA-binding motif, which is encoded by a conserved DNA sequence of about 180 bp called the homeobox. This sequence is particularly associated with the transcriptional regulators of homeotic, or Hox, genes that were found originally in the fruit fly Drosophila, and are involved in orchestrating development in higher eukaryotes. Mutations in animal Hox genes convert one body part into another. Drosophila has two Hox clusters, but vertebrates have four clusters of 9 to 11 genes each on different chromosomes. Vertebrate Hox genes are expressed in different patterns and at specific embryological stages. There is a compelling comparability between these developmental regulators and the HD1/HD2genes of the fungal mating type factors in Schizophyllum and Coprinopsis, as well as Ustilago.
Coprinopsis cinerea and Schizophyllum commune exhibit tetrapolar heterothallism, determined by two mating type factors, called A and B. These are both considered to be MAT loci and the natural population contains many different mating types. In crosses these behave like multiple ‘alleles’ of the two mating type loci. Molecular analysis has revealed that each mating type locus is a very complex region containing several or even many genes (which is why we refer to mating type factors). The genes at A encode transcription factor homeodomain proteins and constitute the HD locus; genes at B make up the P/R locus and encode lipopeptide pheromones and pheromone receptors. Mating type factors are located on different chromosomes, and even conventional genetic analysis has demonstrated internal structure, identifying subloci that are called Aα, Aβ, Bα and Bβ. In S. commune these subloci are relatively far apart, in terms of linkage distance, but they are much closer in Coprinopsis cinerea. The α and β subloci are functionally redundant in the sense that a difference need exist at only one of them for compatibility. Nine versions of Aα, and 32 of Aβ result in 288 different A mating type specificities in S. commune. In Coprinopsis cinerea, there are an estimated 160 A mating type specificities in the natural population. Because there are so many different mating types, most encounters between isolates in nature will be compatible and fertile, and this drives the frequency of outbreeding to greater than 99%. On the other hand, from any one meiosis, say the heterozygote A1B1 × A2B2, because the two MAT loci segregate independently, four different types of progeny are produced (A1B1, A2B2, A1B2, and A2B1) and so these systems are called tetrapolar mating systems.
In saprotrophic basidiomycetes hyphal anastomoses occur readily as part of the maturation process of the mycelium, and anastomosis is independent of the mating type factors. Promiscuous cell-cell fusion can result when two monokaryotic hyphae of Coprinopsis cinerea and other Agaricomycetes encounter each other, irrespective of whether they are compatible at MAT loci (and see Section 4.10). However, for the development and maintenance of the dikaryon, nuclei of interacting partners must carry different allelic versions of genes in at least one sublocus of both P/R and HD loci. A compatible mating in these fungi, which is characterised by clamp connections and conjugate nuclear divisions in the mated hyphae, requires that both A and B are different. In the belief that in this state the mating type factors are fully active, it is called A-on, B-on (Fig. 8). Other than dikaryosis itself, there are some characteristic morphological differences between the monokaryotic and dikaryotic mycelia of Coprinopsis cinerea: branches emerge from monokaryotic hyphae at a wide angle (40 to 90°), but at an acute angle (10 to 45°) from dikaryotic hyphae; monokaryons, but not the dikaryons, produce asexual arthrospores, called oidia, in droplets of fluid; and the aerial mycelium of monokaryons is generally less dense and fluffy than that of dikaryons.
Cytological observations and genetic analyses of partially compatible interactions in Coprinopsis cinerea and Schizophyllum commune have shown that the P/R system (mating type factor B) is involved in events that follow hyphal fusion:
- the reciprocal exchange and migration of nuclei;
- and clamp cell fusion.
On the other hand, genes encoded at the HD locus (mating type factor A):
- repress asexual sporulation;
- regulate pairing of nuclei within the dikaryotic tip cell;
- and coordinate nuclear division, clamp cell formation, and septation from the subapical cell.
Dikaryons arise when both A and B are different but heterokaryons can also be formed in matings in which one of the mating type factors is homozygous. When A factors are the same (called common-A, or A-off, B-on, see Fig. 8), nuclear migration takes place but no clamp connections form. Mating between strains carrying the same B factor forms a heterokaryon (called common-B or A-on, B-off) only where the mated monokaryons meet because nuclear migration is blocked. In this case, conjugate divisions occur, and apical cells of heterokaryotic hyphae start to make clamp connections and the nuclei divide but the clamp (hook) cell fails to fuse with the adjacent cell and the nucleus in the clamp remains trapped (Fig. 8). If the two ‘mates’ share identical alleles at P/R and HD loci, no nuclear exchange occurs, and the mycelial individuals will resume monokaryotic growth.
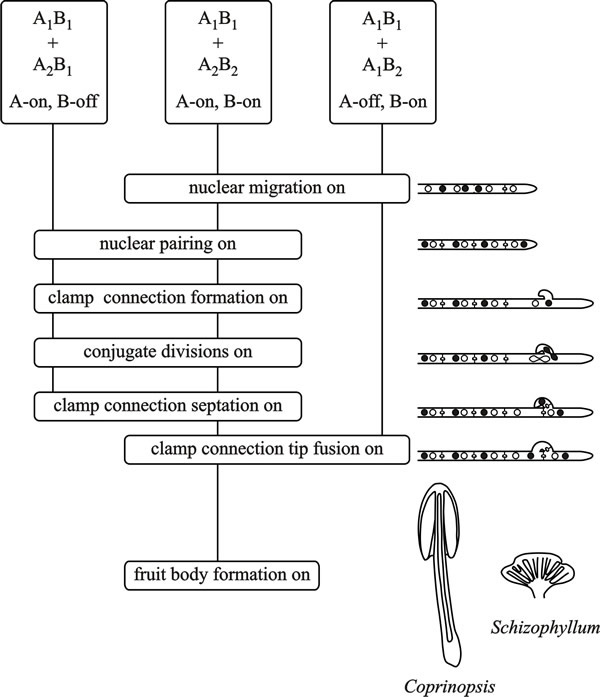 |
| Fig. 8. Flow chart diagram of A (the HD locus) and B (the P/R locus) factor activity in the basidiomycetes Coprinopsis cinerea and Schizophyllum commune. From top to bottom the flow chart depicts the events that take place when two haploid mycelia confront each other. Dikaryon formation requires that both A and B factors are different (which is taken to mean that both A and B functions are turned on (A-on, B-on)). This is depicted in the central vertical line for a confrontation of A1B1 + A2B2, the vertical line connecting ‘function boxes’, the phenotypes of which are indicated in the cartoons on the right hand side. When only A functions are turned on (A-on, B-off), in a common-B heterokaryon (A1B1 + A2B1) conjugate divisions and clamp formation occur, but the clamp connections remain incomplete and without clamp cell fusion nuclear migration cannot take place. In a common-A heterokaryon (A1B1 + A1B2) the A factors are the same but B factors are different (A-off, B-on) and nuclear migration can occur, but without conjugate division or clamp connection formation. Modified from Chapter 2 in Moore & Novak Frazer, 2002. |
This ‘division of labour’ between A and B factors is not universal; nuclear migration is regulated by the B factor alone in Coprinopsis cinerea and Schizophyllum commune, but in Coprinus patouillardii migration is regulated by both factors but the A alone determines fertility. In Ustilago and Tremella, cell fusion is controlled by a locus with two idiomorphs, but the ability of fused cells to grow as dikaryons depends on a second locus with multiple idiomorphs (see the Ustilago section above).
Clearly, the mating type factors determine the initial self/nonself recognition that follows the first anastomoses of the encounter; they also regulate mycelial morphogenesis (up-regulating features that characterise the dikaryon and down-regulating some monokaryotic features, like making oidia) but as it is only dikaryons that fruit in normal circumstances, the A and B mating type factors also regulate fertility.
Nuclear migration through an established mycelium has also been studied in some basidiomycetes. In the species that have been examined in most detail, Coprinopsis cinerea and Schizophyllum commune, basidiospores germinate to form homokaryotic mycelia with uninucleate cells, which is usually called a monokaryon or homokaryon. Two monokaryons will form hyphal anastomoses, and if they are compatible, nuclei will migrate from one mycelium into the other. This establishes a new mycelium, called a dikaryon, which has regularly binucleate cells containing one nucleus of each parental type.
The growth of dikaryotic hyphal tips requires that the two nuclei complete mitosis together (conjugate division) and a mechanism of nuclear migration and sorting that depends on a small backward-growing branch (called a clamp connection or hook cell) at each hyphal septum (side-panel in Fig. 8, above; and see Section 7.4). Nuclei will migrate from a compatible (dikaryotic) mycelium and convert another monokaryon into a dikaryon. In experiments with Coprinopsis radiatus (a very close relative of Coprinopsis cinerea) nuclei invaded mycelia in such circumstances at a rate of 1.5 mm h-1, which is at least four times higher than the hyphal growth rate. In Coprinellus congregatus, a nuclear migration rate of 4 cm h-1 (yes, we do mean centimetres!) has been reported, and migration rates in Schizophyllum commune range up to 2.7 mm h-1 with a hyphal growth rate of only 0.22 mm h-1.
During nuclear migration the invading nucleus undergoes regular division and one of the daughter nuclei moves into the next cell, the intervening dolipore septa between adjacent cells of the monokaryon being broken down into simple pores through which the nuclei can be squeezed. Although some corresponding cytoplasmic movement has been observed in Schizophyllum commune, nuclear migration occurs more commonly without visible cytoplasmic flow.
Nuclear migration is a highly active transport process involving microtubular motors in a manner analogous to the involvement of spindle fibres in the movement of chromosomes during division, and nuclear-identification by mating type factors presumably accounts for its specificity (Debuchy, 1999; Shiu & Glass, 2000) (and see Section 5.12). Clearly, in most cases a specific nuclear type is being transported in a specific direction, and in all this discussion of nuclear migration it is important to emphasise that only nuclei migrate; mitochondria are not exchanged between compatible mycelia. During migratory dikaryotisation, cells lacking nuclei and multinucleate cells are observed, so the dikaryotic state is not set up as soon as compatible cells fuse. Rather, ordered dikaryotic growth emerges after an interval of disorganised and irregular growth.
Detailed molecular analysis of the A mating type factors show that they contain many more genes than classical genetic analysis could reveal (Fig. 9). In fact, each A locus of C. cinerea contains a variable number of genes, which are arranged in pairs like the bE-bW pair in the U. maydis b locus. The C. cinerea gene pairs are designated groups 1, 2, and 3. Each group within the A locus encodes two dissimilar homeodomain proteins (HD1 and HD2) which are homologous to the S. cerevisiae MAT α2 and MAT a1 mating proteins, respectively (Casselton & Olesnicky, 1998; Brown & Casselton, 2001; Kües, 2015; Coelho et al., 2017). .
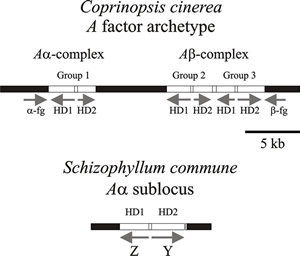 |
Fig. 9. Diagrams of parts of the A (HD locus) mating type factors in Coprinopsis cinerea and Schizophyllum commune. Arrows show the direction of transcription. The (predicted) archetypal A factor from Coprinopsis cinerea has three pairs of functionally redundant genes (group 1, group 2, & group 3) which encode the homeodomain proteins (HD1 & HD2). The α-fg and b-fg sequences are homologous in all A mating type specificities in C. cinerea. Interaction between HD1 and HD2 proteins is the basis of the compatible reaction (see Figs 8 and 10). A factors examined in different strains of C. cinerea isolated from nature contain different combinations, and different numbers, of these genes. In Schizophyllum commune the mating type genes are called Z and Y and encode HD1 and HD2 respectively. Again, different idiomorphs are found in different natural mating types; indeed, the Z gene is absent in the Aα1 mating type. This simplified diagram modified from Chapter 2 in Moore & Novak Frazer, 2002. For much more detailed information about a wider range of fungi refer to Kües, 2015; and Coelho et al., 2017). |
Several features combine to ensure that there is no intragenic recombination
within the A locus, which is likely to disturb its regular ‘two-by-two’
structure. The groups are organised into cassettes so that they act as a single
unit and the DNA sequences are sufficiently different between groups 1, 2 and 3
to avoid homologous recombination. In addition, the paired genes are transcribed
in opposite directions (Fig. 9). The A locus is bounded by DNA
sequences that are homologous in all A mating
type specificities, called α-fg and
b-fg; also, the group 1gene pair is separated from groups 2 and
3 by a 7 kbp DNA sequence that is homologous in all A loci (known as
the ‘homologous hole’). The group 1 gene pair corresponds to the Aα
sublocus defined by conventional linkage analysis, while the group 2 and group 3
gene pairs comprise the Ab
sublocus; 7 kbp is approximately equivalent to the 0.1% recombination observed
between these subloci. These short homologous sequences limit recombination to
the regions between the homeodomain loci.
![]() Basidiomycete sexual development is triggered by a dimerisation between HD1
and HD2 proteins from the different A mating type factors of compatible
individuals (Fig. 10). The N-terminal regions of these proteins are essential
for choosing a compatible partner but not for regulating gene transcription.
Sequences of different idiomorphs of the mating type genes are dissimilar and
have been interpreted as being equivalent to the highly variable region in major
histocompatibility loci in mammals that forms a self/non self recognition
system.
Basidiomycete sexual development is triggered by a dimerisation between HD1
and HD2 proteins from the different A mating type factors of compatible
individuals (Fig. 10). The N-terminal regions of these proteins are essential
for choosing a compatible partner but not for regulating gene transcription.
Sequences of different idiomorphs of the mating type genes are dissimilar and
have been interpreted as being equivalent to the highly variable region in major
histocompatibility loci in mammals that forms a self/non self recognition
system.
The N-terminal region of the mating type protein product is the part that is essential for such self/non self recognition. It ensures that monomers from the same mating type idiomorph are not compatible, and that only the heterodimers made between the products of the two compatible mating type factors present in the cell are able to form the DNA-binding transcription regulators. It has also been shown that compatible protein-protein interactions (heterodimerisation) is more important to compatibility at the A locus than is the occurrence of two homeodomains. The HD2 homeodomain is crucial to DNA binding, but the homeodomains of HD1 proteins are relatively dispensable.
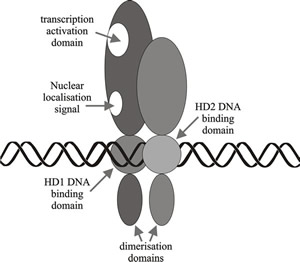 |
Fig. 10. Schematic diagram illustrating a simplified model of homeodomain protein interactions involved in A (HD locus) mating type factor activity in Coprinopsis. Modified from Chapter 2 in Moore & Novak Frazer, 2002. |
The A locus of Schizophyllum commune also controls nuclear pairing, clamp connection formation, conjugate nuclear division and clamp septation. The Aa locus contains the Y gene (which has alleles Y1, Y3, and Y4) and the Z gene (with alleles Z3 and Z4), which encode the homeodomain proteins HD2 and HD1, respectively. The S. commune Aα locus corresponds to a single gene pair from the Coprinopsis cinerea complex. The Ab locus also encodes a polypeptide with a homeodomain. Y and Z are the only determinants of Aα activity, and Aα and Ab function independently of each other. Interactions of the Y and Z proteins have been demonstrated experimentally for non self combinations (for example, Y4 with Z5) proteins, but no interaction occurs between Y and Z proteins encoded by the same A factor (for example Y4 and Z4).
Cloning the B sequences revealed that this mating type factor codes for several pheromone and receptor genes. In the smut fungi, Ustilago, pheromone signalling is important in cell fusion, in establishing the dikaryon, and in maintaining filamentous growth. However, hyphal anastomosis (= hyphal fusion) of monokaryotic vegetative cells in S. commune and C. cinerea does not depend on pheromone-based recognition. In saprotrophic basidiomycetes hyphal anastomoses occur readily as part of the maturation process of the mycelium, and anastomosis is independent of the mating type factors. On the other hand, it is now quite clear that pheromone signalling controls the B-regulated events of reciprocal nuclear migration and clamp cell fusion.
Sequencing the Schizophyllum commune Bα1 region revealed a pheromone receptor gene (called bar1, standing for B-alpha-receptor-1) and three pheromone genes, bap1, bap2 and bap3 (bap stands for B-alpha-pheromone). The Bb1 locus also contains a receptor gene (bbr1) and genes for pheromones, bbp1(1) and bbp1(2). The B factor of Coprinopsis cinerea contains 3 groups (called 1, 2, 3) of genes which each code for a pheromone receptor and two pheromones. The B pheromone genes are all predicted to encode for lipopeptides similar to the Saccharomyces cerevisiae a factor while the B pheromone receptors are homologous to the S. cerevisiae a factor receptor, which is a typical G protein coupled receptor. So far, only the a factor-type pheromones have been found in basidiomycetes whereas both a- and α-type receptors are present (Kothe, 1999).
The model of pheromone function that has been developed for these filamentous basidiomycetes is that after anastomosis, the pheromones produced by the invading nucleus diffuse ahead and act as advance signals of nuclear migration (see Section 5.12). This activates receptors encoded by resident nuclei in nearby cells and the interactions prepare the cells for nuclear migration by initiating septum dissolution to allow nuclei to pass through. A-factor functions then establish the dikaryotic state and clamp connection formation.
Pheromone signalling is then further involved in clamp cell fusion. There are nine mating type specificities at the Bα factor of Schizophyllum commune, each encoding a receptor and one or more pheromones. Hence, each receptor must distinguish at least eight non self pheromones. Individual pheromones may also activate more than one receptor. Work with mutants in B-regulated functions is beginning to identify the genes subject to pheromone signalling. Some of these mutations map to the B loci and affect mating specificity and could be modifiers of pheromone or pheromone receptor gene specificity. Others include nine genes that influence nuclear migration. Many of these genes are linked to B, suggesting that related functions are clustered.
Updated September, 2021
