5.10 mRNA translation and protein sorting
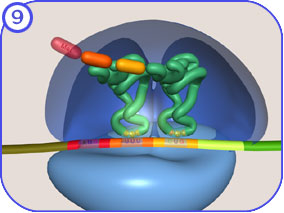
Translation is the synthesis of a protein in the cytoplasm, using an mRNA template that has been exported from the nucleus (Clancy & Brown, 2008). For protein biosynthesis, the mRNA molecules bind to protein-RNA complexes called ribosomes in the cytoplasm, where they are translated into polypeptide sequences. The ribosome is another one of these RNP machines that mediates the formation of a polypeptide sequence based on the mRNA nucleotide sequence (Fig. 4). Initiation of translation begins when the small subunit of the ribosome attaches to the methylated cap at the 5’-end of the mRNA and moves to the translation initiation site. Another key molecule is the family of transfer RNA (tRNA) molecules, which carry anticodons complementary to the mRNA codons. The first codon to be translated in the mRNA is typically AUG (the general translation start signal), and the tRNA that corresponds to this carries the amino acid methionine.
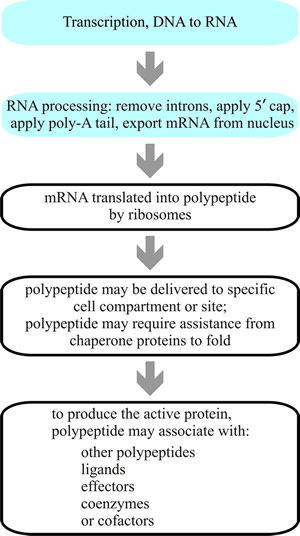 |
Fig. 4. Flowchart giving an overall summary of protein synthesis. Transcription of DNA into RNA takes place within the nucleus (light blue panels; more detail in Figs 2 and 3), and the primary transcript RNA is then modified to add cap and poly-A tail and remove introns. The resulting mature mRNA is exported from the nucleus into the cytoplasm for translation into protein. The mRNA is translated by ribosomes that use tRNA charged with specific amino acids to create the polypeptide corresponding to the sequence of codons carried by the mRNA. Newly-synthesised proteins often carry signal sequences that enable then to be sorted and delivered to specific sites and may be further modified, such as by enzymic activation chemically modifying them, by binding to effectors or coenzymes, and perhaps by binding to other polypeptides. |
The large ribosome subunit now binds to the assembly, creating the peptidyl (or P) site and aminoacyl (or A) sites on the ribosome. The first tRNA occupies the P-site, and the second tRNA, complementary to the second codon, enters the A-site. A peptide bond is made between the methionine (at the P-site) and the second amino acid at the A-site; next, the first tRNA exits, and the ribosome moves along the mRNA to free up the A-site so that the next tRNA can enter. As peptide elongation continues, the growing peptide is always transferred to the tRNA at the A-site by being peptide-bonded to the amino acid carried by that tRNA. The ribosome then moves along the mRNA, so that the tRNA carrying the peptide synthesised to date remains at the same place on the mRNA, held by the hydrogen bonding between its anticodon and the mRNA codon, but is shifted to the ribosome’s P-site by movement of the ribosome.
Note also that amino acids are linked to their tRNA via their carboxy group (= aminoacyl-tRNA), so by definition the first amino acid to be placed in the peptide will have a free amino group, and the last amino acid to be added will have a free carboxylic acid group (once the tRNA is removed). So synthesis is initiated at the amino-terminal end and ends at the carboxy-terminal end of the polypeptide. When a stop codon is encountered in the A-site, a release factor rather than a tRNA enters the A-site and translation is terminated. When termination is reached, the ribosome dissociates, and the newly formed polypeptide is released and folds into a functional three-dimensional protein molecule.
Resources Box CLICK HERE to visit a YouTube animation describing TRANSLATION
This animation belongs to the Virtual Cell Animation Collection produced by the Molecular and Cellular Biology Learning Center at North Dakota State University http://vcell.ndsu.nodak.edu/animations/home.htm
|
The termination of synthesis is not the end of the story. Only a minority of the proteins synthesised in the cytoplasm actually function there. Most proteins have to be delivered to one or other of the cellular organelles to carry out their function, and they may need to be delivered to the inner space of an organelle, to the organelle’s interior membrane(s), to the cell’s outer membrane, or be exported totally to its exterior. This delivery process is carried out using information contained in the protein itself; these proteins have a stretch of amino acid residues in the chain, called signal peptides or targeting peptides, which target the protein to its proper destination.
This addressing mechanism is called the protein targeting or protein sorting mechanism. During transit through the cytoplasm proteins called chaperonins become associated with a newly-synthesised polypeptide to modify its folding and deliver it in a suitable form to the proteins of translocator complexes embedded in the organelle membrane that import it into the organelle. When it is delivered a complex called the signal peptidase removes the signal sequence (so although the coding for this sort of signal sequence must be contained within the mRNA sequence, it does not appear in the final working protein).
|
Resources Box The Nobel Prize in Physiology or Medicine 1999 was awarded to Günter Blobel “...for the discovery that proteins have intrinsic signals that govern their transport and localisation in the cell...” CLICK HERE to visit the Nobel Prize pages at: http://nobelprize.org/nobel_prizes/medicine/laureates/1999/ for more detail of this (needs Internet connection). |
There are two types of targeting peptides: presequences, and internal targeting peptides. Presequences are usually short amino acid sequences at the amino-terminal end of the peptide made up of basic and hydrophobic amino acids. The internal targeting signals are stretches of the primary sequence; to function as signal sequences these stretches have to be brought together on the protein surface by its folding. They are called signal patches.
An amino-terminal signal sequence is the first to be translated and is recognised by a signal recognition particle (SRP) while the polypeptide is still being synthesised (Akopian et al., 2013). SRP binds to the ribosome and targets it to the membrane-bound SRP receptor of the translocon in the membrane of the membranous organelle called the endoplasmic reticulum (ER). When the translating ribosome arrives at the ER membrane the translocon channel is initially closed, but when the signal sequence of the polypeptide is recognised by the channel, it opens, and while it is still being synthesised the protein is inserted into the channel. A junction between ribosome and translocon prevents return of the protein to the cytoplasm.
The signal sequence is immediately removed from the polypeptide by signal peptidase when it is translocated into the ER. Within the ER, the protein is associated with more chaperonin proteins to protect while it folds correctly. Once folded, the protein may be further modified (by glycosylation, for example), before transport to the Golgi apparatus for more processing and transport to other organelles or retention in the ER.
Some proteins are translated in the cytoplasm, independently of the ER, and later transported to their working destination. This happens for proteins intended for the nucleus, mitochondria, chloroplasts in plants, or peroxisomes. Peroxisomes are highly significant to fungi for a variety of reasons (Sibirny, 2016):
- the glyoxylate cycle occurs within the peroxisome so they are necessary for fatty acid metabolism;
- filamentous Ascomycota have specialised peroxisomes called Woronin bodies that plug the septal pore to separate individual cells in a hypha [CLICK HERE to see it now];
- increasing evidence indicates that this organelle can also function as an intracellular signalling compartment and as an organiser of developmental decisions inside the cell;
- peroxisomes contain hydrogen peroxide-generating oxidases and the hydrogen peroxide-detoxifying enzyme catalase.
Interestingly, most peroxisomal matrix proteins have their (type 1) peroxisome-targeting signal sequence at the carboxy-terminal end of the polypeptide. The type 2 (N-terminal) peroxisomal targeting signal occurs in only a few proteins (four are known in humans, one in yeast) (Pollard et al., 2017b).
Some proteins are transmembrane proteins, often transmembrane receptors that pass through their membrane one or more times. These are inserted into the membrane by a translocation process similar to that already described (the first transmembrane domain acts as the first signal sequence) but the process is stopped by an amino acid signal sequence, which is also called the membrane anchor.
The majority of mitochondrial proteins are synthesised in the cytoplasm with appropriate presequences. In this case the ribosome does not dock onto the mitochondrial translocator, but synthesises the preprotein as a cytoplasmic soluble protein. Cytoplasmic chaperonins then deliver the preprotein to a translocator in the mitochondrial membrane.
Resources Box CLICK HERE to visit a YouTube animation describing Mitochondrial Protein Transport
This animation belongs to the Virtual Cell Animation Collection produced by the Molecular and Cellular Biology Learning Center at North Dakota State University http://vcell.ndsu.nodak.edu/animations/home.htm
|
The proteins involved include receptors and the ‘general import pore’ at the outer membrane, which together make up the ‘translocase of outer membrane’ (TOM). The preprotein translocates through TOM in hairpin loops and is then transported through the space between membranes to TIM (‘translocase of inner membrane’) through which it exits to the mitochondrial matrix, where the targeting presequence is removed. Mitochondria are complex organelles and proteins have to be targeted to different parts of them, involving several signals and transport pathways. Targeting to the outer membrane, intermembrane space, and/or inner membrane often requires separate signal sequence(s) in addition to the matrix targeting sequence.
In plants, proteins are also targeted to chloroplasts. We will not dwell on this apart from mentioning that the vast majority of chloroplast proteins are synthesised in precursor form on cytoplasmic ribosomes and are translocated through the TOC (translocon at the outer envelope membrane of chloroplasts) and TIC (translocon at the inner envelope membrane of chloroplasts) complexes. A basic similarity in strategy of mechanism (and naming) is evident between the two major organelles, mitochondria and chloroplasts, which encourages the view that similar mechanisms may be used for other membrane-bound organelles.
For proteins that must get from the cytoplasm to the nucleus, there is a nuclear localisation signal. On the cytoplasmic side of the nuclear membrane, the NPC [CLICK HERE to see it now] recognises proteins carrying a nuclear localisation signal and selectively transports them into the nucleus. Proteins cross the NPC after they have been synthesised and have folded into a three-dimensional shape.
We have to consider protein destruction as the final act in protein sorting. Synthesis might produce a defective protein, a protein may be damaged as it functions, or, more likely, protein function may no longer be required. Such proteins are recycled and are targeted for recycling by ubiquitin (Pickart & Eddins, 2004). Ubiquitin is so called because it is ubiquitous in virtually all types of cells and is one of the most highly conserved (= least changed) proteins. Ubiquitin is a protein consisting of 76 amino acids and in the normal course of events, proteins are inactivated by having several molecules of ubiquitin attached to them; this process is called ubiquitination. Ubiquitin serves as the signal for transport machinery to ferry a protein to a site for proteolysis. There are two pathways of intracellular proteolysis: lysosomal, and proteasomal.
|
Resources Box In 2004, Aaron Ciechanover and Avram Hershko of the Technion Israel Institute of Technology in Haifa, Israel and Irwin Rose of the University of California, Irvine, USA shared the Nobel Prize in Chemistry “...for the discovery of ubiquitin-mediated protein degradation...” CLICK HERE to visit the Nobel Prize pages |
A minority of proteins modified with ubiquitin (in Saccharomyces cerevisiae it tends to be plasma membrane proteins; Hicke, 1999) are internalised and degraded in a lysosome vacuole. The majority of ubiquitin-labelled proteins are transported to the proteasome for degradation. The proteasome is another molecular machine; it’s a cylindrical particle with a central cavity comprising a multicatalytic protease complex together with, at each end, a complex containing several ATPases and a binding site for ubiquitin molecules. The ubiquitinated protein is unfolded by the ATPase complexes, which also recover the ubiquitin and inject the substrate protein into the central chamber where there are several catalytic sites. The substrate protein is cleaved into peptides of between 6 and 9 amino acid residues; the length of the product corresponding to the distance between adjacent catalytic sites in the central chamber of the proteasome.
Updated July, 2019